Abstract: The RInChI is a canonical identifier for reactions which is widely used in reaction databases. It can be used to handle large collections of reactions and to link information from diverse data sources. How much information can it handle? Studies of the SAVI database, which contains more than a […]
InChI Tag: Publication
Can an InChI for Nano Address the Need for a Simplified Representation of Complex Nanomaterials across Experimental and Nanoinformatics Studies? Nanomaterials 2020, 10(12), 2493; https://doi.org/10.3390/nano10122493 Abstract: Chemoinformatics has developed efficient ways of representing chemical structures for small molecules as simple text strings, simplified molecular-input line-entry system (SMILES) and the IUPAC International Chemical Identifier […]
A possible extension to the RInChI as a means of providing machine readable process data Philipp-Maximilian Jacob, Tian Lan, Jonathan M. Goodman & Alexei A. Lapkin Journal of Cheminformatics volume 9, Article number: 23 (2017) Abstract: The algorithmic, large-scale use and analysis of reaction databases such as Reaxys is currently […]
Analysing a billion reactions with the RInChI Jonathan M. Goodman, Gerd Blanke and Hans Kraut From the journal Pure and Applied Chemistry Abstract: The RInChI is a canonical identifier for reactions which is widely used in reaction databases. It can be used to handle large collections of reactions and to […]
International chemical identifier for reactions (RInChI) Guenter Grethe, Jonathan M Goodman & Chad HG Allen Journal of Cheminformatics volume 5, Article number: 45 (2013) Abstract: The IUPAC International Chemical Identifier (InChI) provides a method to generate a unique text descriptor of molecular structures. Building on this work, we report a […]
International chemical identifier for reactions (RInChI) Guenter Grethe, Gerd Blanke, Hans Kraut & Jonathan M. Goodman Journal of Cheminformatics volume 10, Article number: 22 (2018) Abstract: The Reaction InChI (RInChI) extends the idea of the InChI, which provides a unique descriptor of molecular structures, towards reactions. Prototype versions of the […]
Enumeration of Ring – Chain Tautomers Based on SMIRKS Rules Laura Guasch, Markus Sitzmann, and Marc C. Nicklaus https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4170818/ J Chem Inf Model. 2014 Sep 22; 54(9): 2423 – 2432. Abstract: A compound exhibits (prototropic) tautomerism if it can be represented by two or more structures that are related by […]
Tautomerism of Warfarin: Combined Chemoinformatics, Quantum Chemical, and NMR Investigation Laura Guasch, Megan L. Peach, and Marc C. Nicklaus https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7724503/ J Org Chem. 2015 Oct 16; 80(20): 9900-9909. Abstract: Warfarin, an important anticoagulant drug, can exist in solution in 40 distinct tautomeric forms through both prototropic tautomerism and ring chain […]
Experimental and Chemoinformatics Study of Tautomerism in a Database of Commercially Available Screening Samples Laura Guasch, Waruna Yapamudiyansel, Megan L. Peach, James A. Kelley, Joseph J. Barchi, Jr., and Marc C. Nicklaus https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5129033/ J Chem Inf Model. 2016 Nov 28; 56(11): 2149 – 2161. Abstract: We investigated how many cases […]
Tautomerism in large databases Markus Sitzmann, Wolf-Dietrich Ihlenfeldt, and Marc C. Nicklaus https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2886898/ Journal of Computer-Aided Molecular Design volume 24, pages 521-551 (2010) https://link.springer.com/article/10.1007/s10822-010-9346-4 Abstract: We have used the Chemical Structure DataBase (CSDB) of the NCI CADD Group, an aggregated collection of over 150 small-molecule databases totaling 103.5 million […]
Tautomer Database: A Comprehensive Resource for Tautomerism Analyses Devendra K. Dhaked, Laura Guasch, and Marc C. Nicklaus https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8456363/ Abstract: We report a database of tautomeric structures that contains 2819 tautomeric tuples extracted from 171 publications. Each tautomeric entry has been annotated with experimental conditions reported in the respective publication, […]
Toward a Comprehensive Treatment of Tautomerism in Chemoinformatics Including in InChI V2 Devendra K. Dhaked, Wolf-Dietrich Ihlenfeldt, Hitesh Patel, Victorien Delannée, and Marc C. Nicklaus* https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8459712/ Abstract: We have collected 86 different transforms of tautomeric interconversions. Out of those, 54 are for prototropic (non-ring–chain) tautomerism, 21 for ring–chain tautomerism, and 11 […]
Identifying International Chemical Identifier (InChI) Enhancements—QR Codes and Industry Applications IUPAC Project 2015-019-2-800 Chair: Richard M Hartshorn Objective: The International Chemical Identifier (InChI) is a text string that encodes chemical structure and provides a means to search databases for the structure. The InChI Trust <www.inchi-trust.org> is examining development of a […]
Implementation of InChI for chemically modified large biomolecules IUPAC Project 2013-010-1-800 Project chair: Evan Bolton & Keith T. Taylor Objective: To establish requirements and guidelines for the generation of a unique name for biological sequences including chemically modified. The intended outcome is the world-wide adoption of the a standard with […]
IUPAC and the InChI Trust Agree Upon Conditions for Collaboration From the journal Chemistry International — Newsmagazine for IUPAC https://doi.org/10.1515/ci.2010.32.4.16b
IUPAC InChI/InChIKey Project Joins Microsoft BioIT Alliance From the journal Chemistry International https://doi.org/10.1515/ci.2008.30.1.19c
InChI’ng forward: Community Engagement in IUPAC’s Digital Chemical identifier Leah McEwen Chemistry International https://doi.org/10.1515/ci-2018-0109 Chemistry International overview of IUPAC projects related to InChI.
Seminal InChI Publications From the journal Chemistry International https://doi.org/10.1515/ci-2016-0121 2016 overview of some publications on InChI
The InChI Code Paul J. Karol Journal of Chemical Education 2018 95 (6), 911-912 The International Union of Pure and Applied Chemistry has developed a computer algorithmic scheme for unambiguously coding molecular structure into relatively short alphabetic tags. These are now ubiquitous in many descriptions of molecular structure, yet the […]
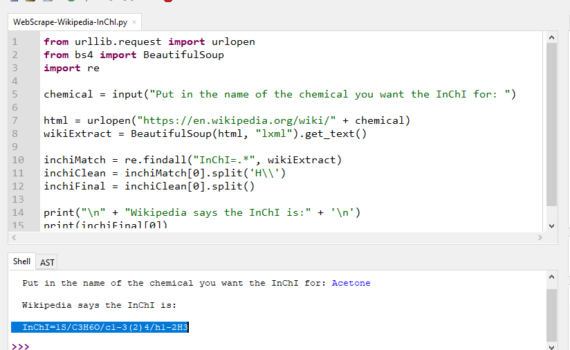
Andrew P. Cornell, Robert E. Belford Chemistry Department, University of Arkansas at Little Rock, Little Rock, Arkansas 72204 Abstract Many individual chemicals have a specific page on Wikipedia that will give information about the use, manufacture and properties of that chemical. The properties that are displayed off to the […]
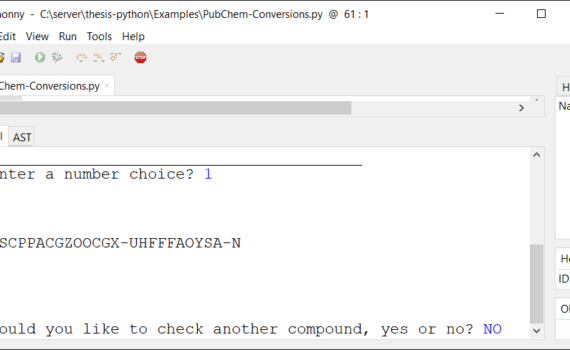
Andrew P. Cornell, Robert E. Belford Chemistry Department, University of Arkansas at Little Rock, Little Rock, Arkansas 72204 Abstract In this tutorial, a program written in Python will take a user specified chemical name and retrieve the associated chemical identifier or basic property using an online chemical database. This […]
PubChem chemical structure standardization Volker D. Hähnke, Sunghwan Kim & Evan E. Bolton Journal of Cheminformatics volume 10, Article number: 36 (2018) Background: PubChem is a chemical information repository, consisting of three primary databases: Substance, Compound, and BioAssay. When individual data contributors submit chemical substance descriptions to Substance, the unique […]
Chemical Entity Semantic Specification: Knowledge representation for efficient semantic cheminformatics and facile data integration Leonid L Chepelev & Michel Dumontier Journal of Cheminformatics volume 3, Article number: 20 (2011 Abstract: Background: Over the past several centuries, chemistry has permeated virtually every facet of human lifestyle, enriching fields as diverse as medicine, agriculture, manufacturing, warfare, and electronics, among […]
Open Data, Open Source and Open Standards in chemistry: The Blue Obelisk five years on Journal of Cheminformatics volume 3, Article number: 37 (2011) Noel M O’Boyle, Rajarshi Guha, Egon L Willighagen, Samuel E Adams, Jonathan Alvarsson, Jean-Claude Bradley, Igor V Filippov, Robert M Hanson, Marcus D Hanwell, Geoffrey R […]
UniChem: a unified chemical structure cross-referencing and identifier tracking system Jon Chambers, Mark Davies, Anna Gaulton, Anne Hersey, Sameer Velankar, Robert Petryszak, Janna Hastings, Louisa Bellis, Shaun McGlinchey & John P Overington Journal of Cheminformatics volume 5, Article number: 3 (2013) Abstract: UniChem is a freely available compound identifier mapping […]
Consistency of systematic chemical identifiers within and between small-molecule databases Saber A Akhondi, Jan A Kors & Sorel Muresan Journal of Cheminformatics volume 4, Article number: 35 (2012) Abstract Background: Correctness of structures and associated metadata within public and commercial chemical databases greatly impacts drug discovery research activities such as […]
Enhancement of the chemical semantic web through the use of InChI identifiers Simon J. Coles,a Nick E. Day,b Peter Murray-Rust,b Henry S. Rzepac and Yong Zhang Org. Biomol. Chem., 2005,3, 1832-1834 Abstract: Molecules, as defined by connectivity specified via the International Chemical Identifier (InChI), are precisely indexed by major web search engines […]
The IUPAC International Chemical Identifier: InChl—A New Standard for Molecular Informatics Alan McNaught CHEMISTRY International November-December 2006
Detection of IUPAC and IUPAC-like chemical names Roman Klinger, Corinna Kolá?ik, Juliane Fluck, Martin Hofmann-Apitius, Christoph M. Friedrich Bioinformatics, Volume 24, Issue 13, July 2008, Pages i268–i276, https://doi.org/10.1093/bioinformatics/btn181 Motivation: Chemical compounds like small signal molecules or other biological active chemical substances are an important entity class in life science publications and patents. Several representations and nomenclatures […]
The IUPAC International Chemical Identifier (InChI) From the journal Chemistry International — Newsmagazine for IUPAC
QSPR modeling of octanol water partition coefficient of platinum complexes by InChI-based optimal descriptors A. A. Toropov, A. P. Toropova & E. Benfenati Journal of Mathematical Chemistry volume 46, pages 1060–1073 (2009 Abstract Comparison of the quantitative structure property relationships (QSPR) based on optimal descriptors calculated with the International Chemical […]
Tautomer Identification and Tautomer Structure Generation Based on the InChI Code Torsten Thalheim, Armin Vollmer, Ralf-Uwe Ebert, Ralph Kühne, and Gerrit Schüürmann Abstract: An algorithm is introduced that enables a fast generation of all possible prototropic tautomers resulting from the mobile H atoms and associated heteroatoms as defined in the […]
How Many Miles Have We Gone, InChI by InChI? by Alex Tropsha and Antony Williams Chemistry International — Newsmagazine for IUPAC https://doi.org/10.1515/ci.2012.34.5.33
Abstract A modified InChI (International Chemical Identifier) string scheme, yaInChI (yet another InChI), is suggested as a method for including the structural information of a given molecule, making it straightforward and more easily readable. The yaInChI theme is applicable for checking the structural identity with higher sensitivity and generating three-dimensional […]
InChIKey collision resistance: an experimental testing Igor Pletnev, Andrey Erin, Alan McNaught, Kirill Blinov, Dmitrii Tchekhovskoi & Steve Heller Journal of Cheminformatics volume 4, Article number: 39 (2012) InChIKey is a 27-character compacted (hashed) version of InChI which is intended for Internet and database searching/indexing and is based on an […]
InChI: connecting and navigating chemistry Antony J Williams Journal of Cheminformatics volume 4, Article number: 33 (2012) Abstracrt: The International Chemical Identifier (InChI) has had a dramatic impact on providing a means by which to deduplicate, validate and link together chemical compounds and related information across databases. Its influence has […]
MMsDusty: an Alternative InChI-Based Tool to Minimize Chemical Redundancy Molecular Informatics, Volume32, Issue8, August 2013, Pages 681-684 Marco Fanton, Matteo Floris, Andrea Cristiani, Stefania Olla, Ricardo Medda, Davide Sabbadin, Alessandro Bulfone, Stefano Moro MMsDusty is an alternative web-oriented InChI-based normalization tool developed with the specific aim to efficiently analyze and remove chemical redundancy, […]