Online mixtures demo, with MInChI generator Cheminformatics 2.0 Blog Post, Alex M. Clark The above blog goes over how to use the MInChI Demo MInChI Demo
InChI Tag: Software
Coordination InChI for inorganics: now with stereochemistry Cheminformatics 2.0 blog by Alex Clark October 18, 2020 Cheminformatics 2.0 Blog Post GitHub: Coordination Complexes for InChI: preliminary study GitHub: Coordination Complexes for InChI: phase 2 study
Tautomerizer – Predict tautomers based on 80+ rules https://cactus.nci.nih.gov/tautomerizer/ Introduction from Web Service (11/24/2022): Experimental service that allows you to test a set of tautomeric transforms with your own molecules. The predefined set of transforms comprises both the current 24 standard rules used by the chemoinformatics toolkit CACTVS and 55+ additional […]
Isotope Enumerator Web GUI isoenum-webgui provides Flask-based web user interface that uses isoenum package to generate accurate InChI (International Chemical Identifier) for NMR metabolite features based on standard NMR experimental descriptions (currently 1D-1H and 1D-CHSQC) in order to improve data reusability of metabolomics data Andrey Smelter, Hunter N.B. Moseley Revision isoenum […]
CAPTURING MIXTURES ” BRINGING INFORMATICS TO THE WORLD OF PRACTICAL CHEMISTRY” Recorded live December 19, 2019 Hosted and presented by the Collaborative Drug Discovery (CDD) Vault Watch our webinar featuring Dr. Chris Jakober (Johns Hopkins), Leah McEwen (Cornell), and Dr. Alex Clark (CDD) to hear about our work toward new […]
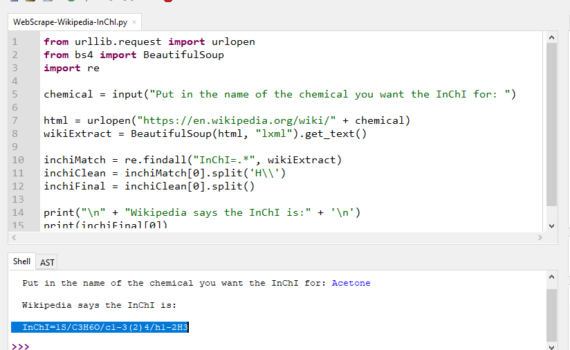
Andrew P. Cornell, Robert E. Belford Chemistry Department, University of Arkansas at Little Rock, Little Rock, Arkansas 72204 Abstract Many individual chemicals have a specific page on Wikipedia that will give information about the use, manufacture and properties of that chemical. The properties that are displayed off to the […]
Andrew P. Cornell, Robert E. Belford Chemistry Department, University of Arkansas at Little Rock, Little Rock, Arkansas 72204 Abstract In this tutorial, a program written in Python will take a user specified chemical name and retrieve the associated chemical identifier or basic property using an online chemical database. This […]
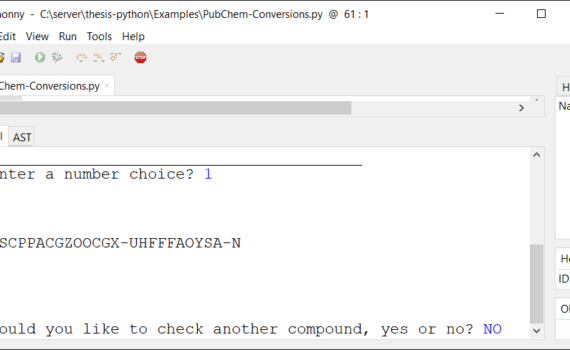
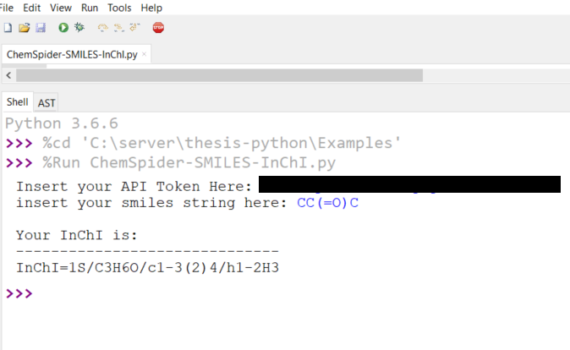
Andrew P. Cornell, Robert E. Belford Chemistry Department, University of Arkansas at Little Rock, Little Rock, Arkansas 72204 Abstract ChemSpider offers many methods in which to access online data through web API (Application Programming Interface) interactions.1 This tutorial will explain how to write a few simple lines of code […]
PubChem chemical structure standardization Volker D. Hähnke, Sunghwan Kim & Evan E. Bolton Journal of Cheminformatics volume 10, Article number: 36 (2018) Background: PubChem is a chemical information repository, consisting of three primary databases: Substance, Compound, and BioAssay. When individual data contributors submit chemical substance descriptions to Substance, the unique […]
Open Data, Open Source and Open Standards in chemistry: The Blue Obelisk five years on Journal of Cheminformatics volume 3, Article number: 37 (2011) Noel M O’Boyle, Rajarshi Guha, Egon L Willighagen, Samuel E Adams, Jonathan Alvarsson, Jean-Claude Bradley, Igor V Filippov, Robert M Hanson, Marcus D Hanwell, Geoffrey R […]
Consistency of systematic chemical identifiers within and between small-molecule databases Saber A Akhondi, Jan A Kors & Sorel Muresan Journal of Cheminformatics volume 4, Article number: 35 (2012) Abstract Background: Correctness of structures and associated metadata within public and commercial chemical databases greatly impacts drug discovery research activities such as […]
MMsDusty: an Alternative InChI-Based Tool to Minimize Chemical Redundancy Molecular Informatics, Volume32, Issue8, August 2013, Pages 681-684 Marco Fanton, Matteo Floris, Andrea Cristiani, Stefania Olla, Ricardo Medda, Davide Sabbadin, Alessandro Bulfone, Stefano Moro MMsDusty is an alternative web-oriented InChI-based normalization tool developed with the specific aim to efficiently analyze and remove chemical redundancy, […]
On InChI and evaluating the quality of cross-reference links Jakub Galgonek & Ji?í Vondrášek Journal of Cheminformatics volume 6, Article number: 15 (2014) Background: There are many databases of small molecules focused on different aspects of research and its applications. Some tasks may require integration of information from various databases. […]
Isotopic (iso) enumerator (enum) – enumerates isotopically resolved InChI (International Chemical Identifier) for metabolites. The isoenum Python package provides command-line interface that allows you to enumerate the possible isotopically-resolved InChI from one of the Chemical Table file (CTfile) formats (i.e. molfile, SDfile) used to describe chemical molecules and reactions as […]
This RDKit InChI Calculation with Jupyter Notebook tutorial is useful to teach the basics of how to interact with InChI using a cheminformatics toolkit in a Jupyter Notebook. The notebook has the following learning objectives: Setup RDKit with a Jupyter Notebook Construct a molecule (RDKit molecular object) from a SMILES […]
Spjuth et al. Journal of Cheminformatics 2013, 5:14 Abstract Background: The InChI algorithms are written in C++ and not available as Java library. Integration into software written in Java therefore requires a bridge between C and Java libraries, provided by the Java Native Interface (JNI) technology. Results: We here describe […]